Flex-Seq
Flex-Seq™ is a high-throughput targeted genotyping platform for commercial next generation sequencing applications. Focusing on scalability paired with data accuracy, reproducibility, and completeness allows Flex-Seq Ex-L to deliver industry-scale solutions for industry-scale genotyping
A genotyping league of its own
Flex-Seq is a high-throughput targeted genotyping platform for commercial next generation sequencing applications. Focusing on scalability paired with data accuracy, reproducibility, and completeness allows Flex-Seq to deliver industry-scale solutions for industry-scale genotyping.
Flex-Seq genotype data matches chip-based genotyping array technology, ensuring consistency between legacy datasets. Genotyping data from other technologies can also be incorporated into Flex-Seq assays and custom/novel genotyping markers can be developed for any species.

Flex-Seq at a glance
Full-service pipeline
- Tissue/DNA to SNPs
Greater balance and specificity than multiplexed PCR
Target tens of thousands of markers
- Novel marker panels
- Legacy data compatible
- Industry standard panels
Used for:
- Genomic selection and parentage testing
- Plant and animal breeding
- Population genetics
- Pathogen detection and analysis
Resources
FAST
- Up to 2-week turnaround time – tissue/DNA to SNPs
FOCUSED
- Up to 98% marker recovery
- Up to 99% on-target sequencing
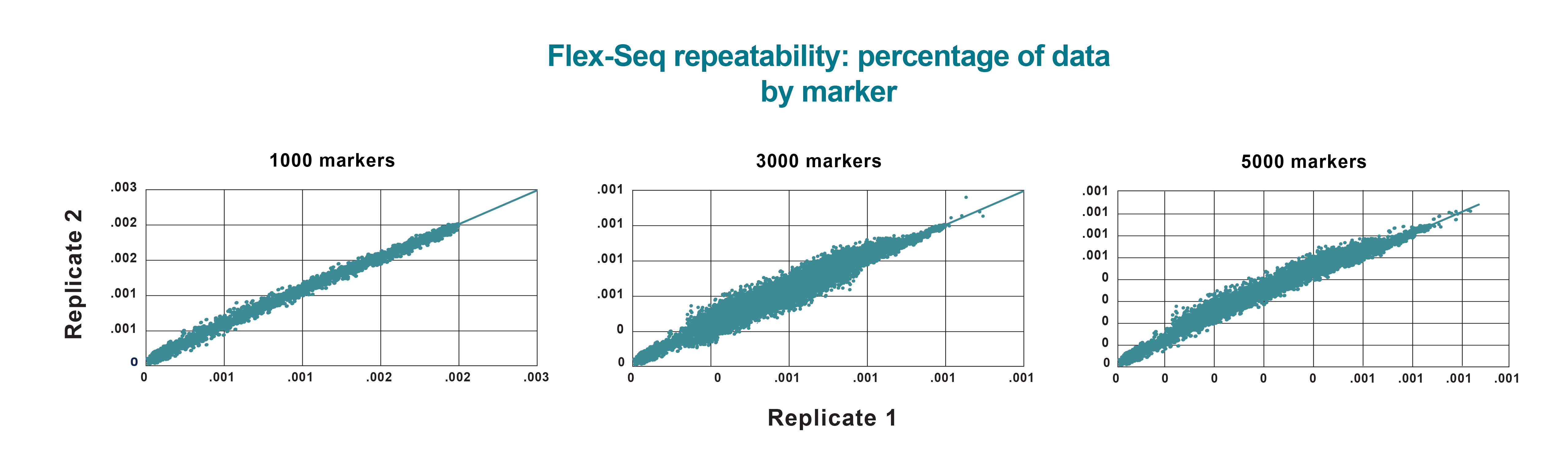
- 99% agreement between technical reps
- 98% agreement with array technology
FLEXIBLE
- Update Flex-Seq panels by adding/removing markers for evolving needs
- Low DNA input
For SNP discovery and characterising hundreds of thousands of targets of any species (including complex polyploids) check out Capture-Seq.
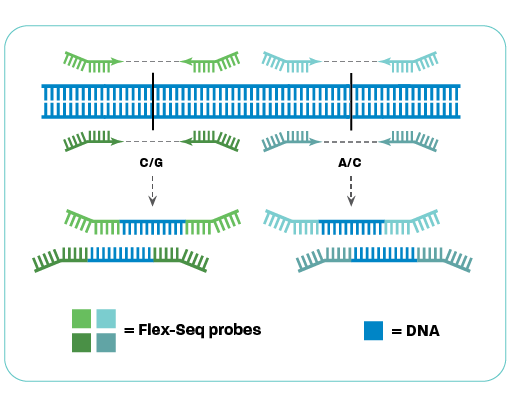
Flex-Seq technology
Multiplexed hybridisation > PCR
The secret is Flex-Seq probe technology
- Higher multiplexing with cleaner data than multiplexed PCR
- Higher marker recovery than multiplexed PCR
- Greater flexibility to include targets than chip-based arrays
SNPs within a 300 bp window may be suitable for haplotyping. Phased alleles improve breeding selections, especially in polyploid species.
Flex-Seq targets individual regions using two Flex-Seq probes, often focusing on a single SNP. Probes can be designed to flexibly bind on either side of the SNP, leading to greater ability to recover desired targets. Once both probes bind, the complementary sequence is synthesised between the probes and the target region is amplified from the genomic DNA. The genomic DNA and non-specific binding products are removed. The proprietary Flex-Seq probes enable high levels of target multiplexing in a single reaction.
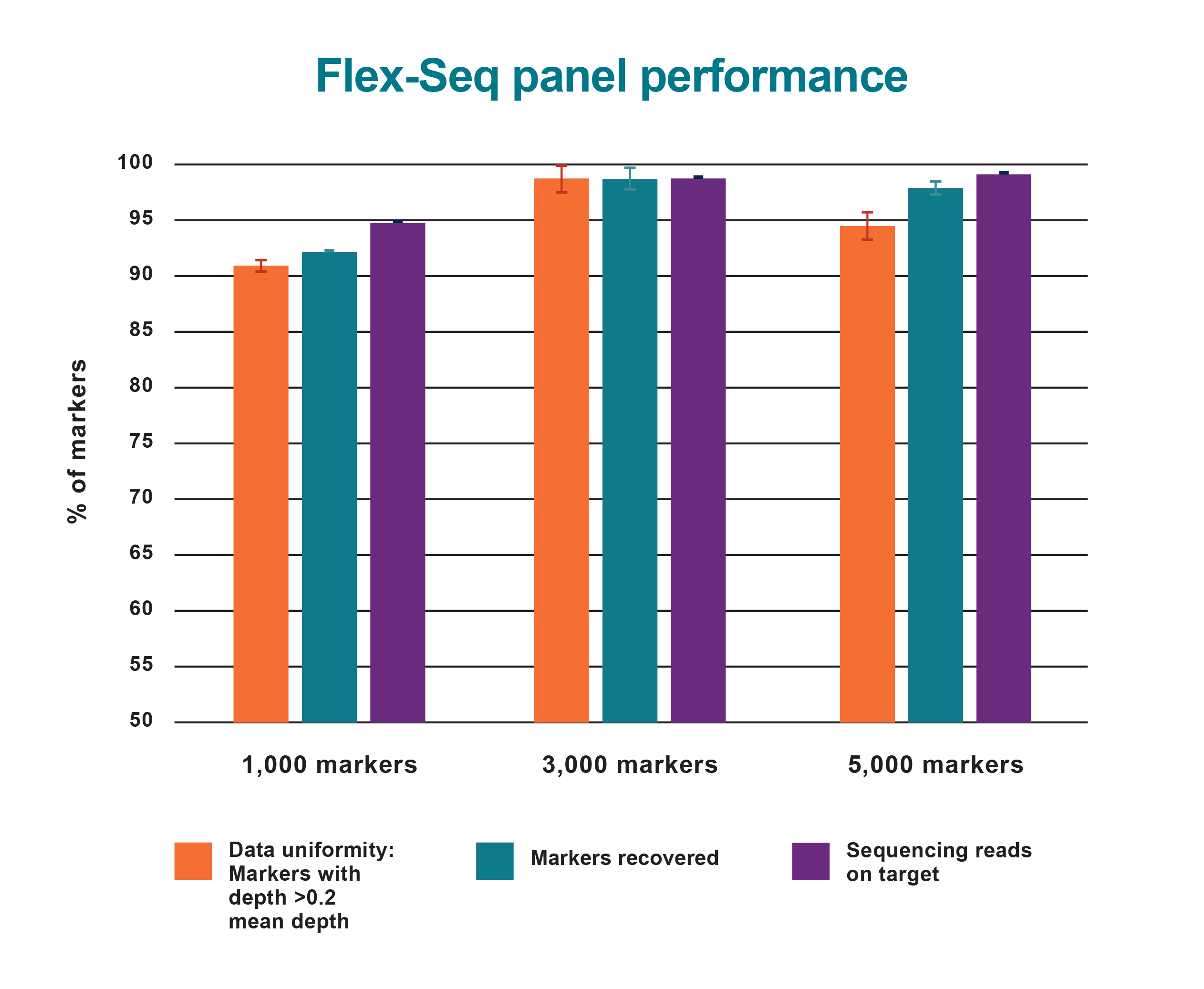
Flex-Seq panel performance
Summary metrics of genotyping results from three Flex-Seq marker panels.
Data Uniformity displays the percentage of markers with >0.2x mean sequence depth (91%-98%).
Markers Recovered shows the percentage of markers (92%-98%) with sufficient sequencing depth for accurate genotyping (i.e. 10X per haploid genome).
Sequencing reads on Target reflects the reaction specificity, with 95-99% of all aligned sequencing data mapping to the intended targets.
The combination of high marker recovery, data uniformity and reaction specificity ensures efficiency and overall completeness of genotyping results for any marker panel.
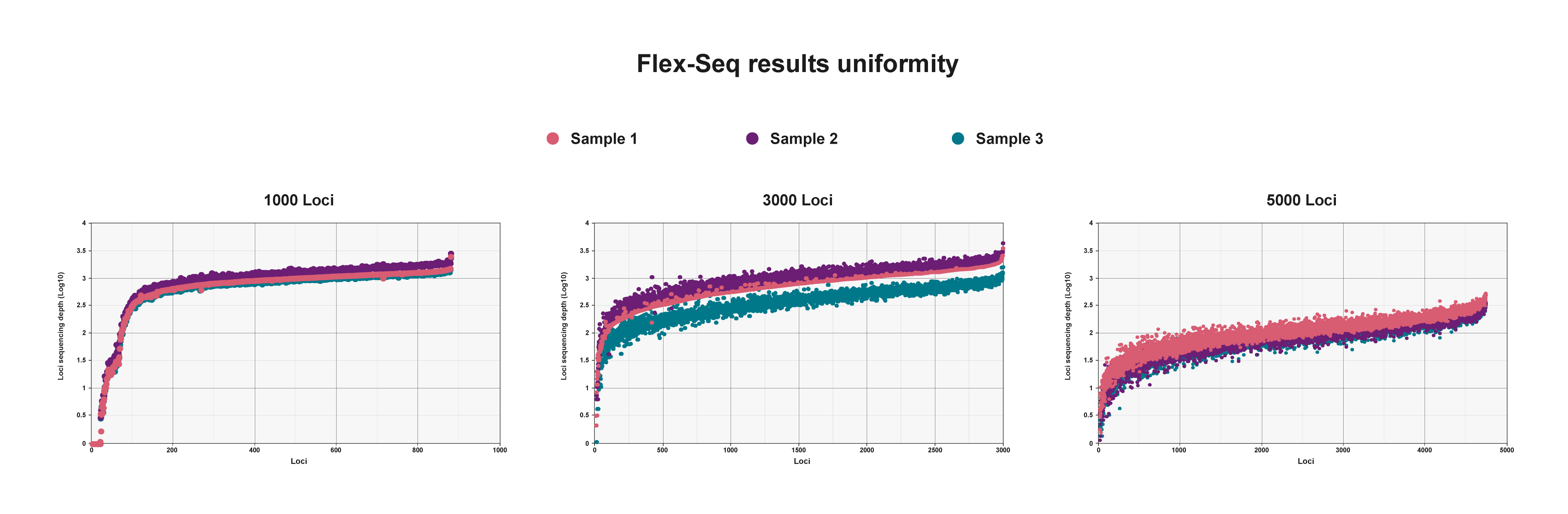
Flex-Seq results uniformity
Log transformation of sequencing depth per marker using three different marker panels. Uniform sequencing depth across all markers enables efficient multiplexed sequencing of thousands of samples.
These efficiencies can be replicated in novel marker panel designs to enable customised targeting of thousands of markers across the genome.
Flex-Seq results reliability
Correlation of the percentage of reads generated per marker between technical replicates using three different marker panels. Flex-Seq demonstrates high levels of consistency in marker recovery and data quantity per marker from independent sample reactions. High repeatability translates to a more predictable recovery of markers from sample to sample, improving the ease of additional data analysis and completeness of final results.