Capture-Seq
Capture-Seq is a targeted next generation sequencing (NGS) platform for characterising hundreds of thousands of targets (including complex polyploids). Samples may be comprised of a single species, a mix of closely related species or distantly related taxa. Capture-Seq is suitable for both high and low volume applications. There is no minimum sample number required to utilise an existing marker assay or for custom assay design.
Finding needles in the genomic haystack
Capture-Seq is the most comprehensive targeted genotyping system for discovering and characterising the genome for:
- Allele mining: Pre-breeding marker discovery
- SNP genotyping and haplotyping
- QTL mapping and candidate gene identification
- Genetic fingerprinting
- Polyploid genomic selection
- Resistance gene sequencing (RenSeq)
- Pan-genome construction and mapping
- Any organism can be analysed using Capture-Seq, and we offer a suite of validated, ready-made Capture-Seq panels for industry-wide standardisation of genotyping data.

Resources
Targeted exome sequencing
- Hundreds of thousands of targets
- Predesigned and custom panels available
- Legacy data compatible
- Short and long read options
- Illumina®
- PacBio®
Tissue/DNA to Fastq data, SNPs and more
- Minimum 500 samples for DNA extraction
- Up to 5-week turnaround times
Repeatable and reliable
- Sequence the same regions every time
- <1% missing target data
Suitable for commercial and research agriculture
- Low minimum sample number
- Minimal ascertainment bias
Capture-Seq vs arrays and GBS
| Genotyping system | Targeted genotyping data | Data interval per target | Development cost | Ease of customisation and adaptation | Missing data % | Ascertainment bias | Read depth at targets (equivalent sequencing) | Repeatability | Suitable for novel marker discovery | Suitable for haplotype reconstruction | Ease if copy number and dosage detection | Analytical complexity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Capture-Seq | Yes | 400-500 bp | Low | High | Low | Low | Predictable and controllable | High | Yes | Yes | High | Low |
| Chip-based arrays | Yes | 1 bp | High | Low | Low | High | N/A | High | No | No | Low | Low |
Capture-seq technology
Answering multiple questions in tandem
Capture-Seq flexibly targets SNPs, genes, and QTLs at the same time to produce sequence data at each region, instead of a single SNP, through the power of NGS. This enables Capture-Seq to provide greater genomic insights into breeding material and training populations, including the ability to phase alleles into haplotypes for more effective polyploid results.
Easy and adaptive
Capture-Seq panels are suitable for different breeding programs of the same or closely related species, ensuring comprehensive pre-breeding marker discovery with minimal risk of ascertainment bias and wasted resources.
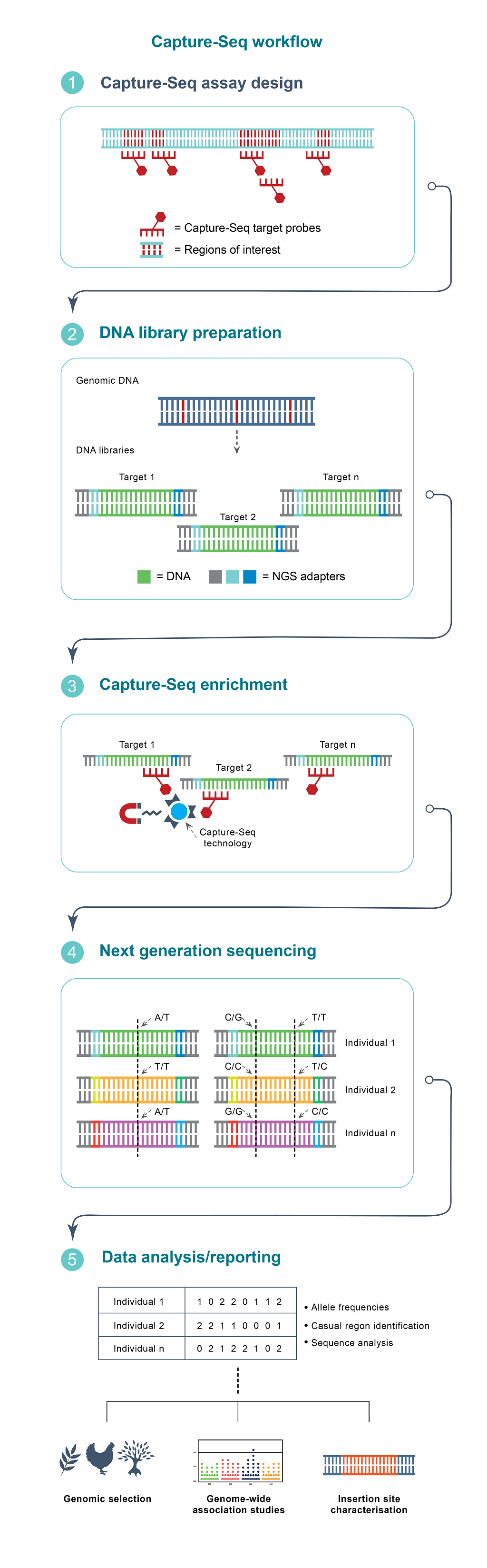
Capture-seq technology diagram (at the right)
Capture-Seq uses targeted probe hybridisation technology to selectively recover regions of interest throughout the genome of any species.
- Capture-Seq probe targets are selected from reference sequence data including genomes and/or transcriptomes. After target selection, Biosearch Technologies designs and synthesises complementary probe sequences to enrich for target regions during sample processing.
- Genomic DNA is processed to sequencing-compatible DNA libraries.
- After DNA library preparation, Capture-Seq probes are hybridised to each sample to selectively enrich target regions over other non-targeted DNA sequences. This improves the proportion of sequencing data going towards regions of interest vs the overall genome.
- After next generation sequencing (NGS), sample data from the target regions is analysed and markers are identified across all samples.
- Markers can be further analysed for use in genomic selection, genome-wide association studies, insertion site characterisations and more.
Capture-seq genotyping results
| Species | Genotyping objective | # Capture-Seq targets | # SNPs detected | Missing data % |
|---|---|---|---|---|
| Maize | Fine mapping resistance genes (R-genes) | 5,000 | 4,623 | 0.15 |
| Sugarcane | Genomic selection and genetic mapping | 10,000 | 37,976 | 0.1 |
| Loblolly pine | Genomic selection | 20,000 | 67,525 | 0.002 |
| Blueberry | Genomic selection and genetic mapping | 31,000 | 205,057 | 0.03 |