How KASP genotyping works
Scientists around the world have been using the KASP™ genotyping technology to transform commercial agriculture for more than two decades, making food production more sustainable and improving crop yields. This proprietary technology enables fast and reliable genotyping of large to small numbers of samples of any species.
The KASP genotyping technology is based on competitive allele-specific PCR and enable bi-allelic scoring of single nucleotide polymorphisms (SNPs) and insertions and deletions (Indels) at specific loci. KASP has been cited in thousands of peer reviewed scientific publications for hundreds of species with millions of KASP markers identified.

The wonderful simplicity of the KASP genotyping technology
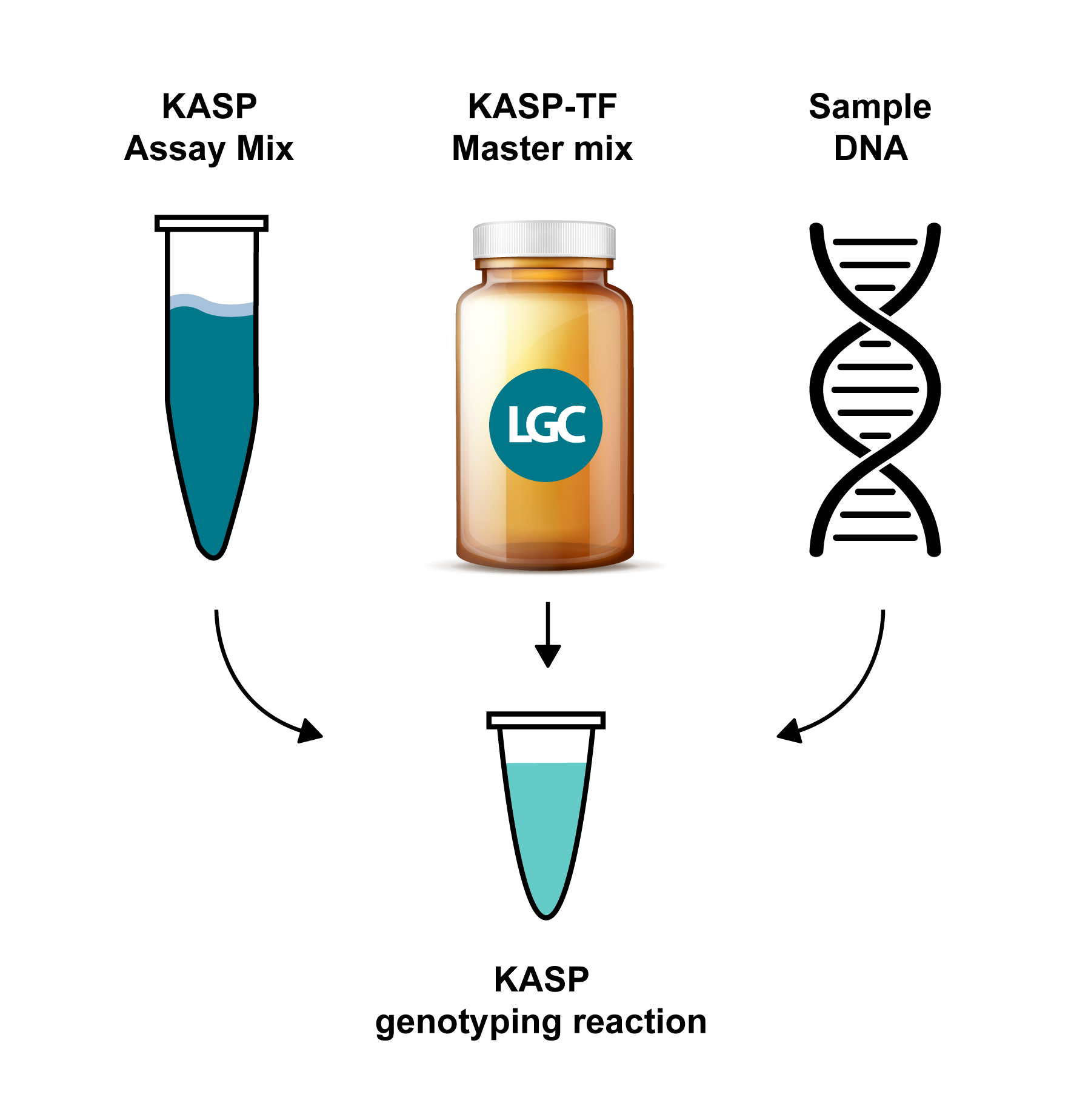
A KASP genotyping reaction has three components:
- SNP-specific KASP assay targeting primers (KASP Assay Mix)
- Universal KASP-TF Master Mix
- Template DNA
Simply combine the three components, run the genotyping assay using a thermal cycler and complete an end-point fluorescent read. KASP delivers > 99.8% accuracy and >90% SNP and Indel assay conversion rate. Completed KASP reactions are readable on most FRET-capable plate readers and qPCR instruments.
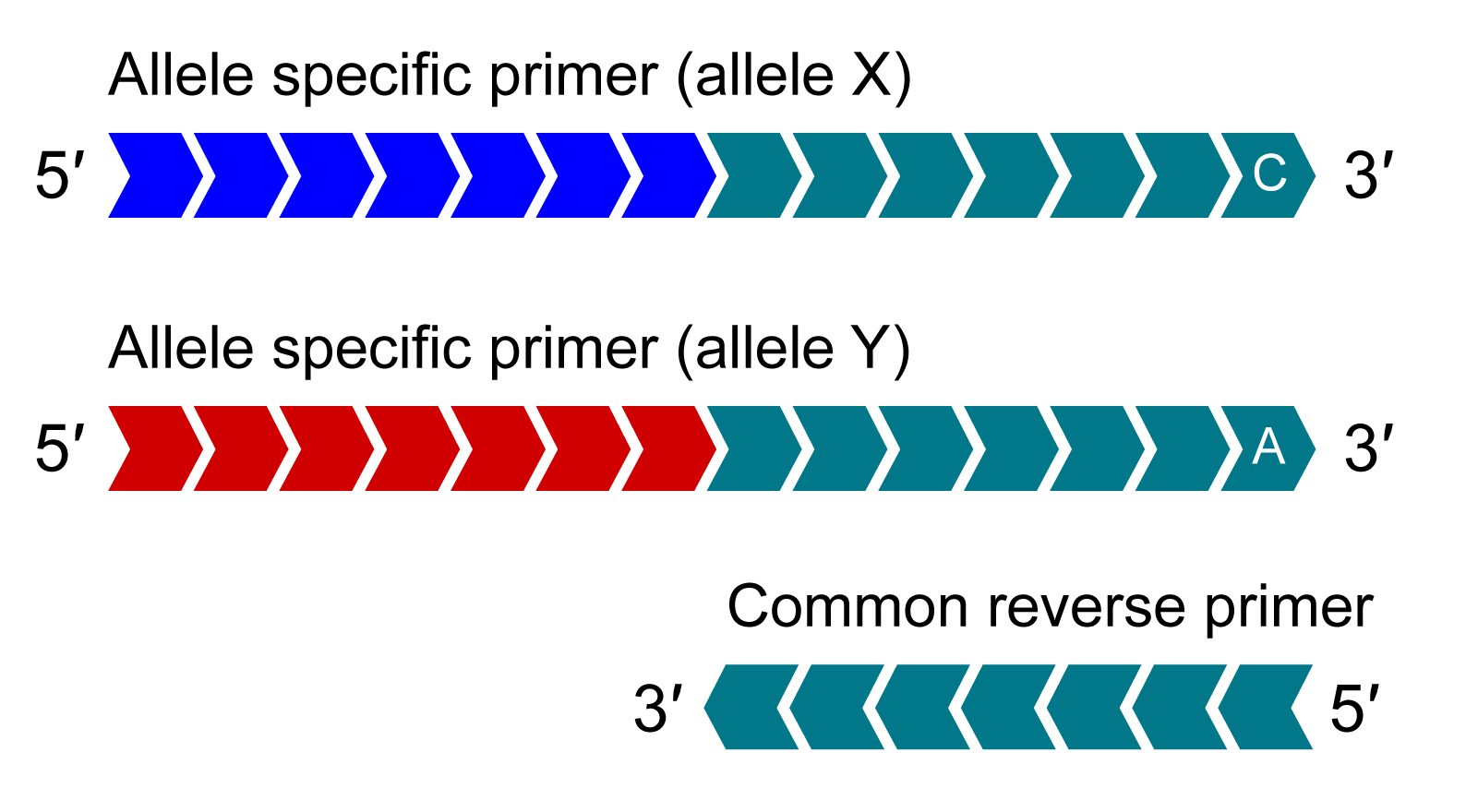
KASP Assay Mix
KASP Assay Mix contains two different allele-specific competing forward primers with unique tail sequences and one common reverse primer.
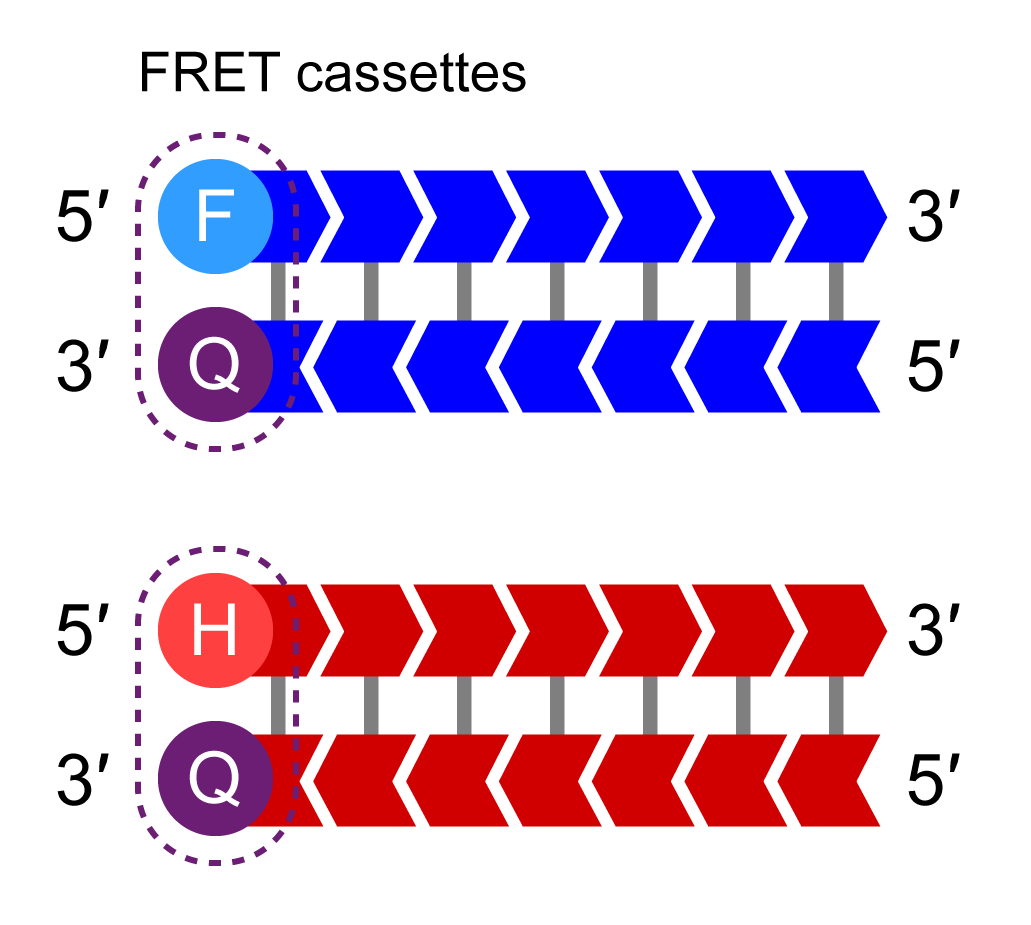
KASP Master Mix
KASP Master Mix contains the FRET cassettes and Taq DNA polymerase in an optimised buffer solution.

Learn more about the components of a KASP genotyping reaction
KASP genotyping process
Denatured template and annealing of primer(s)
In the first round of PCR, the allele-specific primer (or primers if the sample is heterozygous) that matches the target sequence amplifies the target region in conjunction with the common reverse primer. For this illustration, the allele-Y primer binds and is extended.
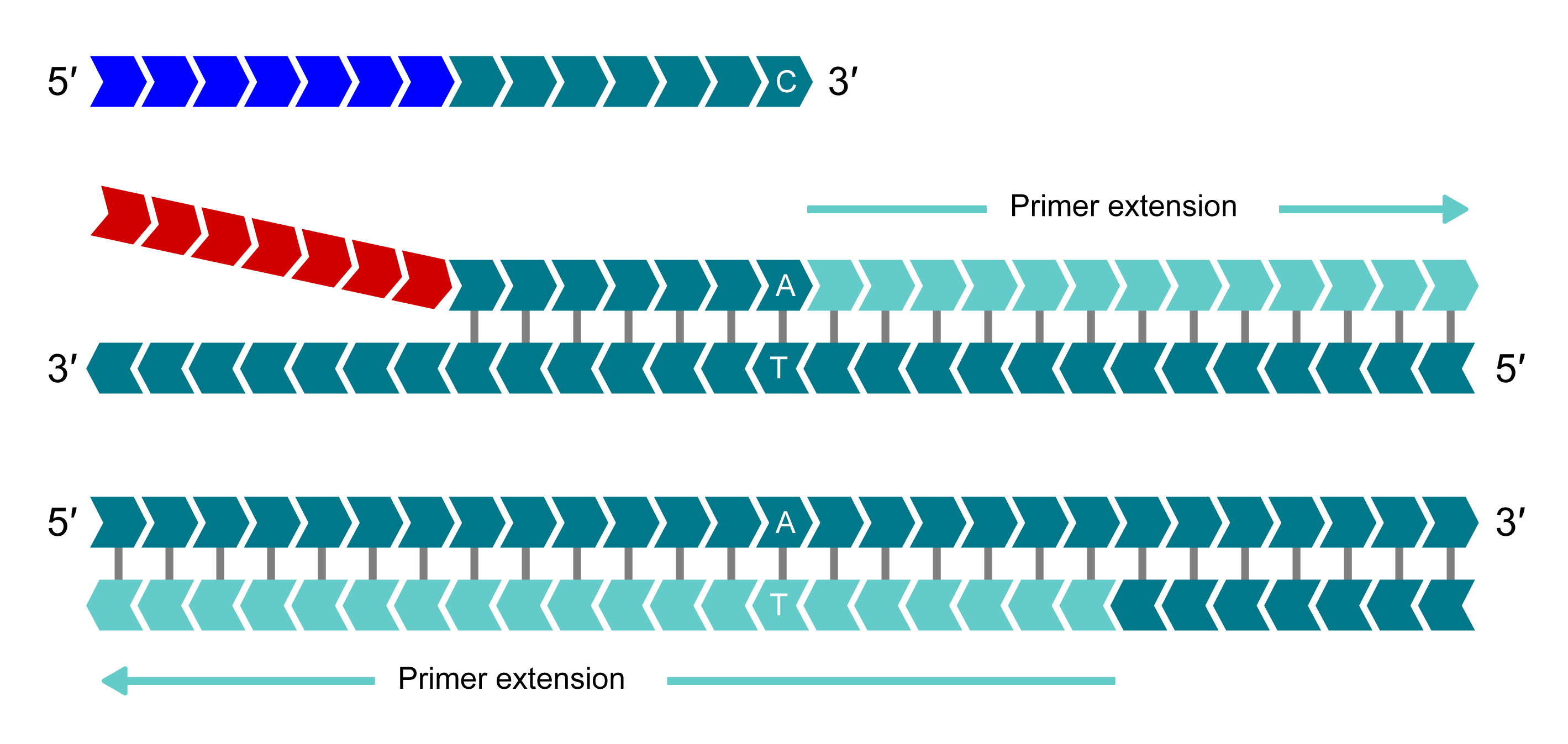
Complement of allele-specific tail sequence generated
In the second round of PCR, the reverse primer binds and is extended, making a complementary copy of the allele-Y tail.

Signal generation
The HEX-labelled oligo binds to the new complementary tail sequence and is no longer quenched.
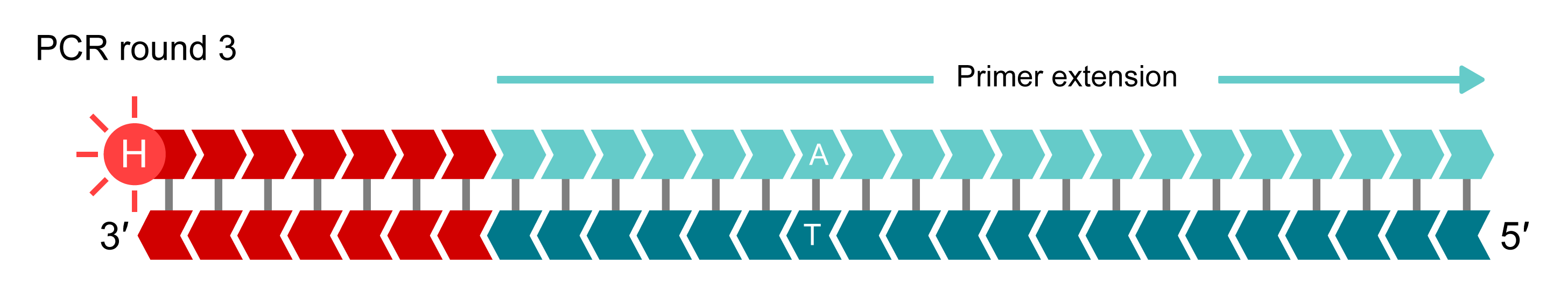
In additional rounds of PCR, levels of allele-specific tail sequence increase. The fluorophore-labelled part of the FRET cassette is complementary to new tail sequences and binds to these sequences, releasing the fluorophore from the quencher to generate a fluorescent signal. In this example, allele-Y (HEX) signal is generated and the fluorophore for the non-incorporated X-allele (FAM) remains quenched.
Bi-allelic discrimination is achieved through the competitive binding of the two allele-specific forward primers. If the genotype at a given SNP is homozygous, only one of the two possible fluorescent signals will be generated. If the genotype is heterozygous, a mixed fluorescent signal will be generated.
Watch this 9-minute video to learn more about the KASP genotyping process
Our scientific experts in KASP genotyping are available to discuss your requirements and work with you to achieve your genotyping goals whether in your lab or with our lab services.
Contact us todayWhy choose KASP for your genotyping assay?
Unmatched quality, reliability, and credibility have established KASP as a foundational SNP genotyping chemistry for agrigenomics. KASP is easy to use – simply add your SNP-specific KASP Assay Mix and the universal KASP-TF Master Mix to your DNA samples, run a thermal cycling program, and measure the end-point fluorescence.
