Genotyping refers to the process of determining genetic variations among individuals in a population. Single nucleotide polymorphisms (SNPs) are the most common type of genetic variation and by definition are single-base differences at a specific locus that is found in more than 1% of the population. SNPs can be found in both coding and non-coding regions of the genome and can lead to phenotypic variations when found in coding regions. These genotype changes can confer positive or negative phenotypic outcomes, such as more robust stress tolerance in crops or enhance the susceptibility for disease in humans. Hence, SNPs are often useful markers for understanding the biology of organisms.
When found in non-coding regions, SNPs act as markers for evolutionary genomics studies. Related to SNPs are “indels”, short for insertions and deletions of nucleotides of varying length. As with SNPs, indels in coding regions can change the amino acid sequence of a protein, either by adding or subtracting an amino acid to the sequence if the indel is in multiples of 3, or by creating a frameshift mutation if the indel is not in a multiple of 3.
A third type of genetic variation is copy number variation (CNV), which results from having different numbers of copies of a DNA segment in various genomes. In cases where the copy number variation is for an encoded gene, the variation can lead to susceptibility or resistance to disease. Some phenotypes are also dosage-sensitive, and the copy number is responsible for shades of variability among members of a species.
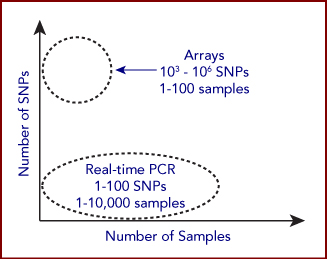
For both SNP genotyping, many methods exist to determine genotype among individuals. The chosen method generally depends on the throughput needs, which is a function of both the number of individuals being genotyped and the number of genotypes being tested for each individual. The chosen method also depends on the amount of sample material available from each individual or sample, which dictates the required sensitivity of the assay.