Licence-free MGB Probes for any application
Minor Groove Binder (MGB) Probes are dual-labeled 5' hydrolysis probes consisting of a 5' fluorescent reporter dye and a 3' Eclipse Dark Quencher (EDQ) conjugated to a MGB moiety. Biosearch Technologies offers high quality yet cost effective MGB oligos with a competitive turnaround time and no minimum order quantity.
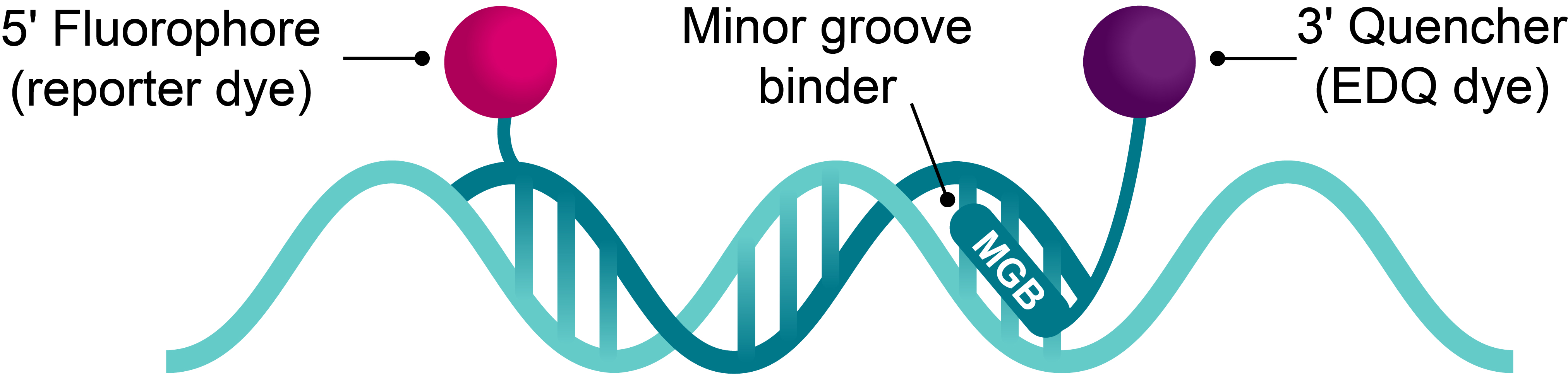
Short probe design delivers high specificity and signal-to-noise
The 3' MGB moiety binds non-covalently to the minor groove and stabilises the target-probe duplex, effectively increasing the Tm of the duplex. The improved stability increases the specificity to the target and allows for a shorter probe design.
A shorter probe has greater quenching efficiency because the dye is closer to the quencher. Combined with a non-fluorescent quencher, such as EDQ, the short MGB Probe leads to low background and high signal-to-noise.
Key benefits
- Short probes with high specificity: The MGB moiety at the 3' end increases the melting temperature (Tm) and stabilises binding, allowing a shorter probe with improved sequence specificity.
- High signal-to-noise for increased sensitivity: The increased quenching efficiency of MGB Probes improves sensitivity with lower background noise and a higher signal-to-noise ratio.
- Improved SNP detection: High duplex stability leads to enhanced ∆Tm and improved SNP/mismatch discrimination.
- Trusted oligo supplier: Biosearch Technologies is one of the most tenured oligo houses. We are known for quality, consistent delivery and flexibility in format.
- ISO 9001 and ISO 13485 manufacturing options: Experience freedom from the constraints and restrictions of your current oligo supplier by partnering with us from assay design to scale up and commercialisation.
Improved SNP/mismatch discrimination
Single base mismatches have a greater destabilising effect on MGB Probes than other probe types, particularly when the mismatches are within the minor groove binding region. This greatly enhances the ∆Tm for a mismatch and improves SNP discrimination.
Trusted supplier of high quality oligos
Our multi-site oligo manufacturing operations provide manufacturing redundancy, risk mitigation and surge capacity. With our unique vertical integration, our chemical manufacturing labs produce most of the critical raw materials used in our own oligo synthesis methods, ensuring a steady and secure supply chain as well as consistent quality at every stage of development. By manufacturing our own dyes, quenchers, specialty amidites, CPG and specialty synthesis columns, we ensure that we are a reliable partner from project inception through commercial scale up.
Manufacturing options under ISO 9001 and ISO 13485
We meet your regulatory needs from research and development to scale up and commercialisation with ISO 9001 (RUO grade) and ISO 13485/cGMP (diagnostic grade) certified manufacturing options, as applicable.
Dye-quencher options
| 5' fluorescent dye | Abs (nm) | Em (nm) | 3' quencher |
|---|---|---|---|
| FAM | 495 | 520 | EDQ* |
| TET | 521 | 536 | EDQ |
| CIV-550 | 530 | 550 | EDQ |
| HEX | 535 | 556 | EDQ |
*Eclipse Dark Quencher (EDQ)
Equivalent performance to industry standard
MGB Probes manufactured at Biosearch Technologies demonstrate equivalent performance to industry-standard MGB Probes.
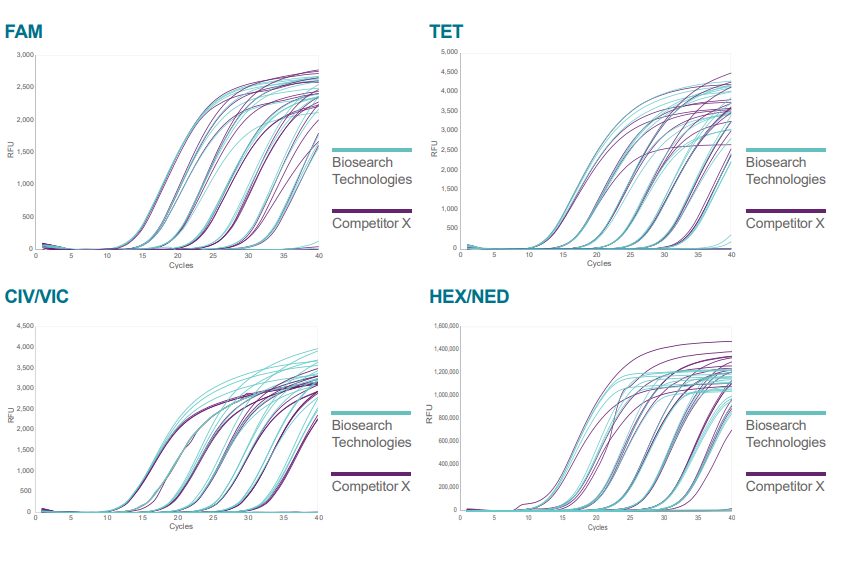 Figure 1. MGB assays were designed for the E. coli gene lacZ. The probes for each assay were synthesised 5’ FAM, TET, CIV, VIC, HEX or NED and 3’ MGB – Eclipse Dark Quencher (EDQ), by both Biosearch Technologies (blue) and Competitor X (purple). All non-labelled oligonucleotides were synthesised by Biosearch Technologies. The performance of each assay was monitored by running a dilution series (1 x 102 to 1 x 108 copies per reaction) of E. coli genomic DNA in TaqMan™ Fast Advanced Master Mix, following manufacturers recommendations for assay concentrations and cycling conditions. Assays were run on a BIORAD CFX96 (FAM, TET and CIV/VIC) or an ABI QuantStudio5 (HEX/NED). The Cq values for the Biosearch Technologies and Competitor X manufactured assays were compared for each dilution point and shown to have equivalent performance, with PCR efficiency and R2 values passing PCR criteria (efficiency 90-110% and R2 > 0.98) for all dyes.
Figure 1. MGB assays were designed for the E. coli gene lacZ. The probes for each assay were synthesised 5’ FAM, TET, CIV, VIC, HEX or NED and 3’ MGB – Eclipse Dark Quencher (EDQ), by both Biosearch Technologies (blue) and Competitor X (purple). All non-labelled oligonucleotides were synthesised by Biosearch Technologies. The performance of each assay was monitored by running a dilution series (1 x 102 to 1 x 108 copies per reaction) of E. coli genomic DNA in TaqMan™ Fast Advanced Master Mix, following manufacturers recommendations for assay concentrations and cycling conditions. Assays were run on a BIORAD CFX96 (FAM, TET and CIV/VIC) or an ABI QuantStudio5 (HEX/NED). The Cq values for the Biosearch Technologies and Competitor X manufactured assays were compared for each dilution point and shown to have equivalent performance, with PCR efficiency and R2 values passing PCR criteria (efficiency 90-110% and R2 > 0.98) for all dyes.
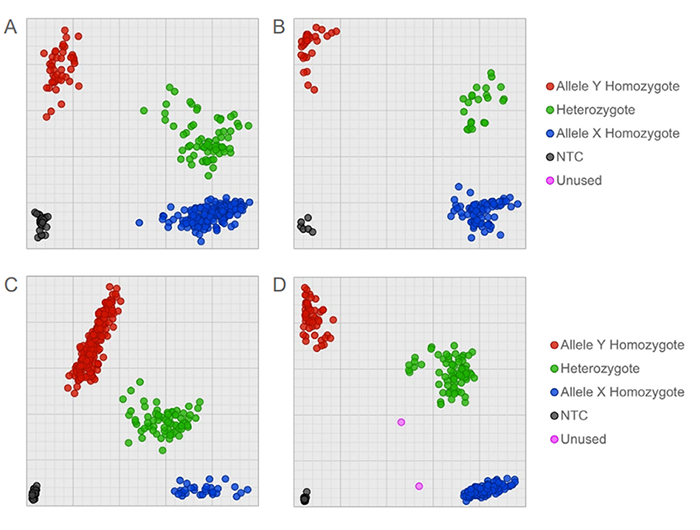
Figure 2. MGB assays synthesised with 5’ FAM or CIV and 3’ MGB – Eclipse Dark Quencher (EDQ) were designed for maize SNP identification. The cluster plots were generated using maize assays: (A) and (B): PZA03069_4; (C): PZA01688_3; (D): PZA02890_4. Full details of all published assays tested are available on the maize genotyping panel Biosearch Technologies webpage. Assays were run on two different Biosearch Technologies low-volume, high-throughput automation platforms, allowing for liquid handling, thermal cycling and fluorescence detection/analysis. Assays were run on the IntelliQube in 1.6 µL reaction volumes in Array Tape (A and B) and the SNPline workflow in 1 µL reaction volumes in 1536-well plates (C and D). Both crude-extracted (A, C and D) and sbeadex-purified (B) DNA gave clear clusters, allowing for clear and easy genotype identification. All assays were run with Biosearch Technologies’ BHQ Probe Master Mix.