Fanzor is a eukaryotic programmable RNA-guided endonuclease
Saito, M., Xu, P., Faure, G., Maguire, S., Kannan, S., Altae-Tran, H., Vo, S., Desimone, A., Macrae, R. K., & Zhang, F.
RNA-guided systems, which use complementarity between a guide RNA and target nucleic acid sequences for recognition of genetic elements, have a central role in biological processes in both prokaryotes and eukaryotes.
Interstrand crosslinking of homologous repair template DNA enhances gene editing in human cells
Ghasemi, H. I., Bacal, J., Yoon, A. C., Tavasoli, K. U., Cruz, C. I., Vu, J. T., Gardner, B. M., & Richardson, C. D.
We describe a strategy to boost the efficiency of gene editing via homology-directed repair (HDR) by covalently modifying the template DNA with interstrand crosslinks.
Targeting TBK1 to overcome resistance to cancer immunotherapy
Sun et al. (2023)
Here we identify the innate immune kinase TANK-binding kinase 1 (TBK1) as a candidate immune-evasion gene in a pooled genetic screen. Using a suite of genetic and pharmacological tools across multiple experimental model systems, we confirm a role for TBK1 as an immune-evasion gene.
Drag-and-drop genome insertion of large sequences without double-strand DNA cleavage using CRISPR-directed integrases
Yarnall et al. (2022)
We present programmable addition via site-specific targeting elements (PASTE), which uses a CRISPR–Cas9 nickase fused to both a reverse transcriptase and serine integrase for targeted genomic recruitment and integration of desired payloads.
Real-Time Culture-Independent Microbial Profiling Onboard the International Space Station Using Nanopore Sequencing
Stahl-Rommel, S., Jain, M., Nguyen, H. N., Arnold, R. R., Auñón-Chancellor, S. M., Sharp, G. M., Castro, C. L., John, K. K., Juul, S., Turner, D. J., Stoddart, D., Paten, B., Akeson, M., Burton, A. S., & Castro-Wallace, S. L.
Here, we report on the development, validation, and implementation of a swab-to-sequencer method that provides a culture-independent solution to real-time microbial profiling onboard the ISS.
Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing
Joung et al. (2020)
Here, we describe a simple test for detection of SARS-CoV-2. The sensitivity of this test is similar to that of reverse-transcription–quantitative polymerase-chain-reaction (RT-qPCR) assays. STOP (SHERLOCK testing in one pot) is a streamlined assay that combines simplified extraction of viral RNA with isothermal amplification and CRISPR-mediated detection.
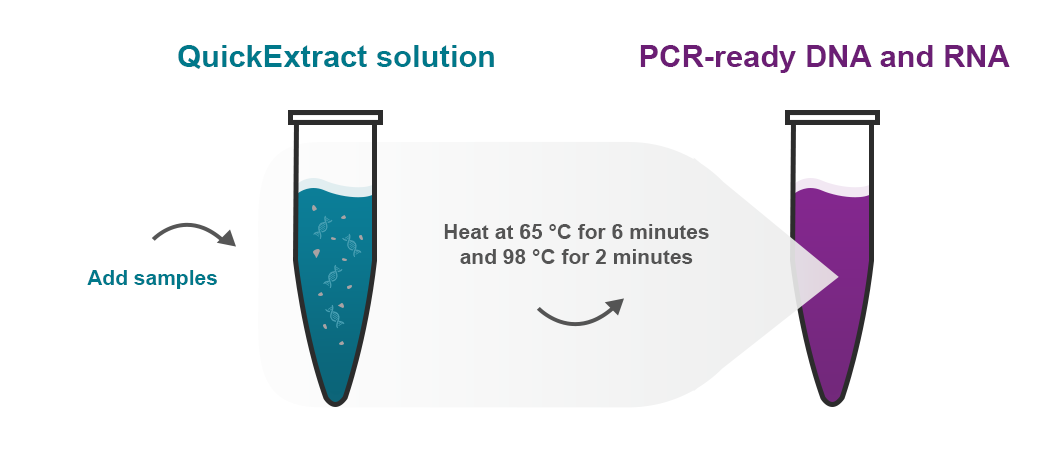
A 5-min RNA preparation method for COVID-19 detection with RT-qPCR
Ladha, A., Joung, J., Abudayyeh, O. O., Gootenberg, J. S., & Zhang, F.
RNA extraction has become a bottleneck for detection of COVID-19, in part because of reagent shortages. We present here a rapid protocol that circumvents the need for RNA extraction that is compatible with RT-qPCR-based detection methods.
Genome engineering using the CRISPR-Cas9 system
Ran, F. A., Hsu, P., Wright, J., Agarwala, V., Scott, D., & Zhang, F.
Here we describe a set of tools for Cas9-mediated genome editing via nonhomologous end joining (NHEJ) or homology-directed repair (HDR) in mammalian cells, as well as generation of modified cell lines for downstream functional studies.